MY PROJECTS
DIGITAL HEALTH HACKATHON 2021
Hosted by: Seneca College
How can open-registry clinical data be used in patient care decision-making for rare diseases?
On the quest to answer this question, me and my teammates envisioned an information tool which could analyze data within an Electronic Health Record (EHR) that provided evidenced based clinical guidelines. We knew that small patient population leads to limited data and greater uncertainty in optimizing patient care, despite ongoing fundamental research in this area.
Decission - Our solution
We designed our clinical decision support tool (Decissio) that could optimize the path of patient's recovery for personalized treatment delivery. Using Cystic Fibrosis as our model disease, our dashboard would assimilate reliable information from open registries and multidisciplinary bioinformatic tools, to enable the creation of a personalized treatment and monitoring plan by clinicians.
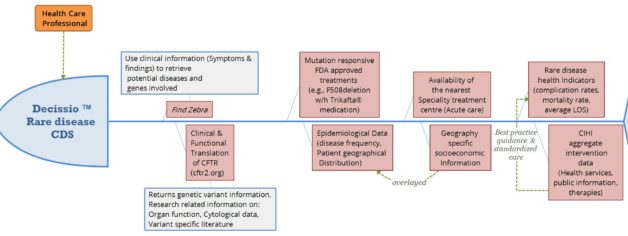

Competition results
Over a short time, we analyzed the data set for linkages between diagnosis & prognosis, developed an innovative solution & presented to an independent panel of judges. We modeled clinician interaction & potential patient outcomes using Tableau and Python. Our solution was voted #1 for:
❖ Best in category for level of innovation
❖ Feasibility and quality of our demonstration
❖ Creation of a prototype system & effective team communication
Watch our short 5-minute presentation to learn more:

(iGEM) International Genetically Engineered Machine Competition
Hosted by: Massachusetts Institute of Technology (MIT) 2015
How do you use standardized genetic parts to solve real-world problems?
Undergraduate students around the world are presented this question each year and compete against other teams in a multidisciplinary setting. My UofT Team and I chose to design a bacteria that could treat tar sands by breaking down petrochemical pollutants. I knew I had to 'teach' it's DNA to recognize & digest pollutants & survive in the process. Now I was no aquaman, and yet we succeeded in our goal. Let me tell you how...
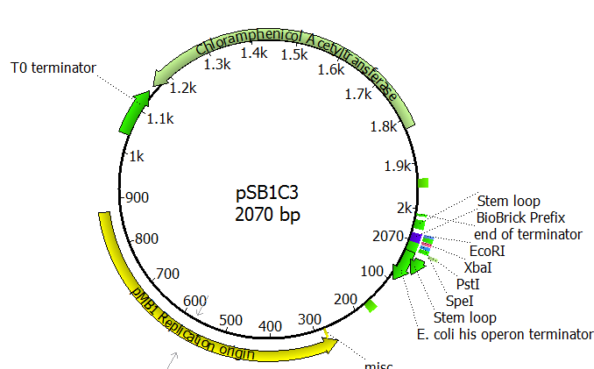
Designer DNA
Using iGEM's library, we ordered the right DNA sequences for enzymes that were upto this monumental task. What came next was the toughest part! Using 'molecular scissors', I developed lab protocols to cut and introduce our designer DNA into E. Coli, culture and simulate our tests in conditions representative of tar sands. Now Imagine doing this about 20 times and we were ready. But we were far from finished! To win over the judges I had to take my team a little further...

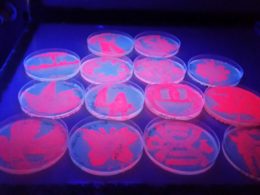
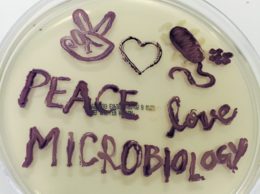
Public Outreach
I had the wonderful opportunity to inspire scientific interest among high school students and foster their passion for life sciences. I thoroughly enjoyed sharing my love for biotechnology over a 5 day hands-on workshop and prepared student's for their journey into university life-science programs. I knew I could spark their interest if I organized micro-biology experiments and have them design their own 'Designer DNA'. We cultured some 'fluorescent bacteria' that you see on the left. It turned out to be a fascinating experience for all. Following it's success, my team was ready to present the hardwork of our 5 month long passion project.
iGEM Results
With our synthetic DNA prepared, we presented at MIT with the hope of pushing the boundaries of synthetic biology and tackling the important issue of environmental pollution. The judges also liked our web-based application that modeled addition of our synthetic E. coli into the Athabasca river microbiome.
The novel DNA parts we created, secured us a silver medal in the Best Applied Design category. It was a challenging and valuable learning experience!

